Numerical simulation#
In this notebook, we’ll go over some simple example use-cases of SBMLtoODEjax to load SBML files from the BioModels website, convert them to jax modules, and run the simulation (with the provided initial conditions).
Imports#
import jax
jax.config.update("jax_platform_name", "cpu")
import matplotlib.pyplot as plt
from sbmltoodejax.utils import load_biomodel
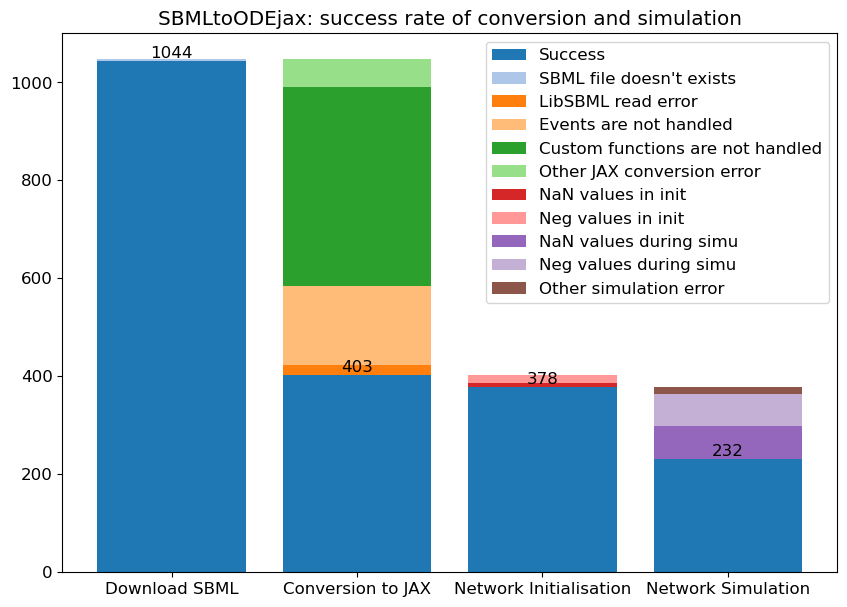
BioMD 10#
See https://www.ebi.ac.uk/biomodels/BIOMD0000000010#Curation
# load and simulate model
model, _, _, _ = load_biomodel(10)
n_secs = 150*60
n_steps = int(n_secs / model.deltaT)
ys, ws, ts = model(n_steps)
# plot time course simulation as in original paper
y_indexes = model.modelstepfunc.y_indexes
plt.figure(figsize=(6, 4))
plt.plot(ts/60, ys[y_indexes["MAPK"]], color="lawngreen", label="MAPK")
plt.plot(ts/60, ys[y_indexes["MAPK_PP"]], color="blue", label="MAPK-PP")
plt.xlim([0,150])
plt.ylim([0,300])
plt.xlabel("Reaction time (mins)")
plt.ylabel("Concentration")
plt.legend()
plt.show()
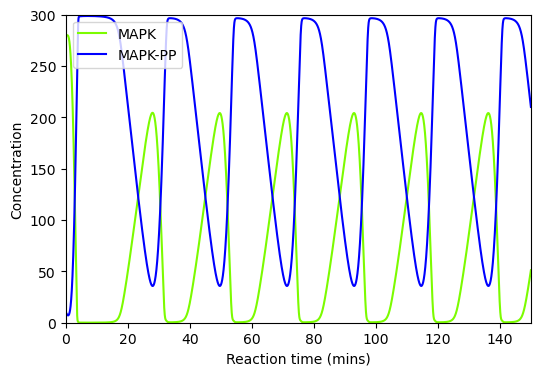
BioMD 37#
See https://www.ebi.ac.uk/biomodels/BIOMD0000000037#Curation
# load and simulate model
model, _, _, _ = load_biomodel(37)
n_secs = 25
n_steps = int(n_secs / model.deltaT)
ys, ws, ts = model(n_steps)
# plot time course simulation as in original paper
y_indexes = model.modelstepfunc.y_indexes
plt.figure(figsize=(6, 4))
plt.plot(ts, ys[y_indexes["S"]], color="red", label="S")
plt.xlabel("Reaction time")
plt.ylabel("Concentration")
plt.xlim([0, 25])
plt.ylim([0, 60])
plt.legend()
plt.show()
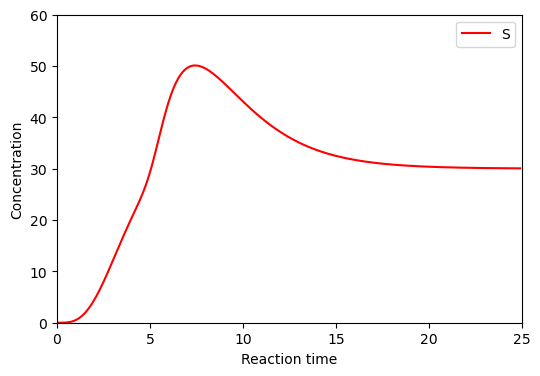
BioMD 50#
See https://www.ebi.ac.uk/biomodels/BIOMD0000000050#Curation
# load and simulate model
model, _, _, _ = load_biomodel(50)
n_secs = 200
n_steps = int(n_secs / model.deltaT)
ys, ws, ts = model(n_steps)
# plot time course simulation as in original paper
y_indexes = model.modelstepfunc.y_indexes
fig, ax = plt.subplots(3, 2, figsize=(12, 10))
ax[0,0].plot(ts, ys[y_indexes["DFG"]], color="red", label="DFG")
ax[0,0].plot(ts, ys[y_indexes["Gly"]], color="lawngreen", label="Gly")
ax[0,0].plot(ts, ys[y_indexes["MG"]], color="blue", label="MG")
ax[0,0].set_ylim([0,12])
ax[0,1].plot(ts, ys[y_indexes["Glu"]], color="red", label="Glu")
ax[0,1].plot(ts, ys[y_indexes["Man"]], color="lawngreen", label="Man")
ax[0,1].plot(ts, ys[y_indexes["Fru"]], color="blue", label="Fru")
ax[0,1].set_ylim([0,.8])
ax[1,0].plot(ts, ys[y_indexes["AA"]], color="red", label="AA")
ax[1,0].plot(ts, ys[y_indexes["FA"]], color="lawngreen", label="FA")
ax[1,0].set_ylim([0,6])
ax[1,1].plot(ts, ys[y_indexes["_3DG"]], color="red", label="3-DG")
ax[1,1].plot(ts, ys[y_indexes["_1DG"]], color="lawngreen", label="1-DG")
ax[1,1].set_ylim([0,.1])
ax[2,0].plot(ts, ys[y_indexes["Mel"]], color="red", label="Mel")
ax[2,0].set_ylim([0,3])
for i in range(3):
for j in range(2):
if (i, j) != (2, 1):
ax[i,j].set_xlim([0,200])
ax[i,j].set_xlabel("Reaction time (mins)")
ax[i,j].set_ylabel("Concentration")
ax[i,j].legend()
else:
ax[i,j].axis("off")
plt.show()
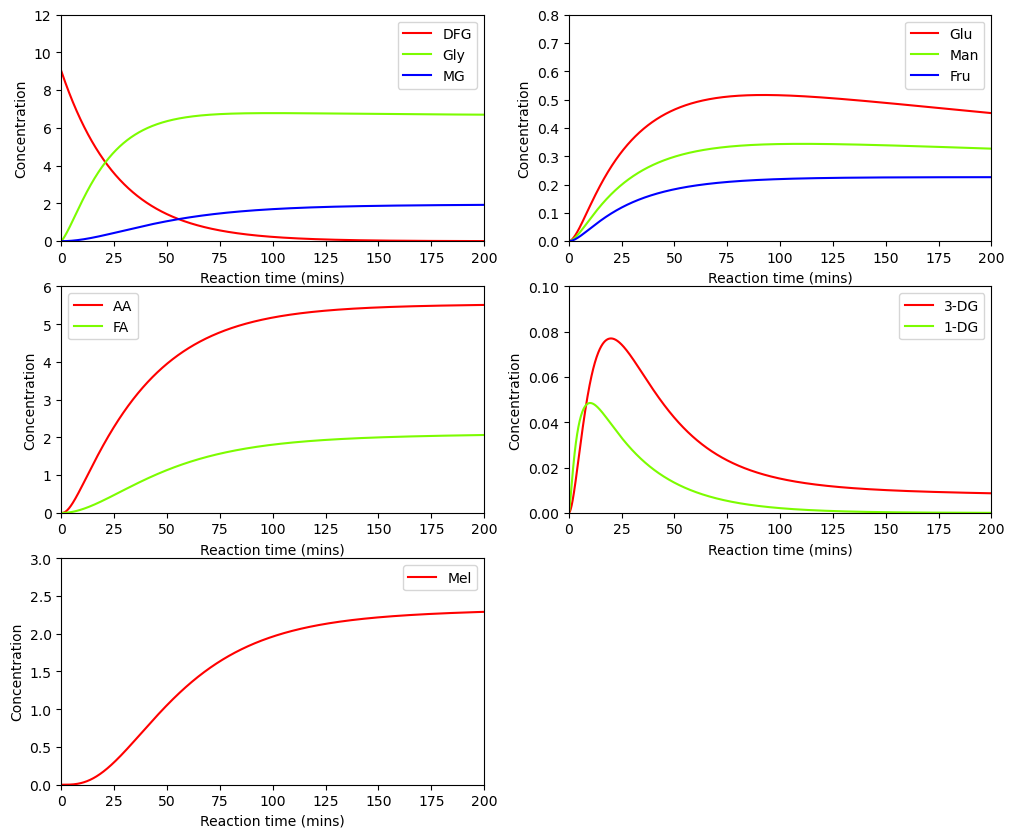
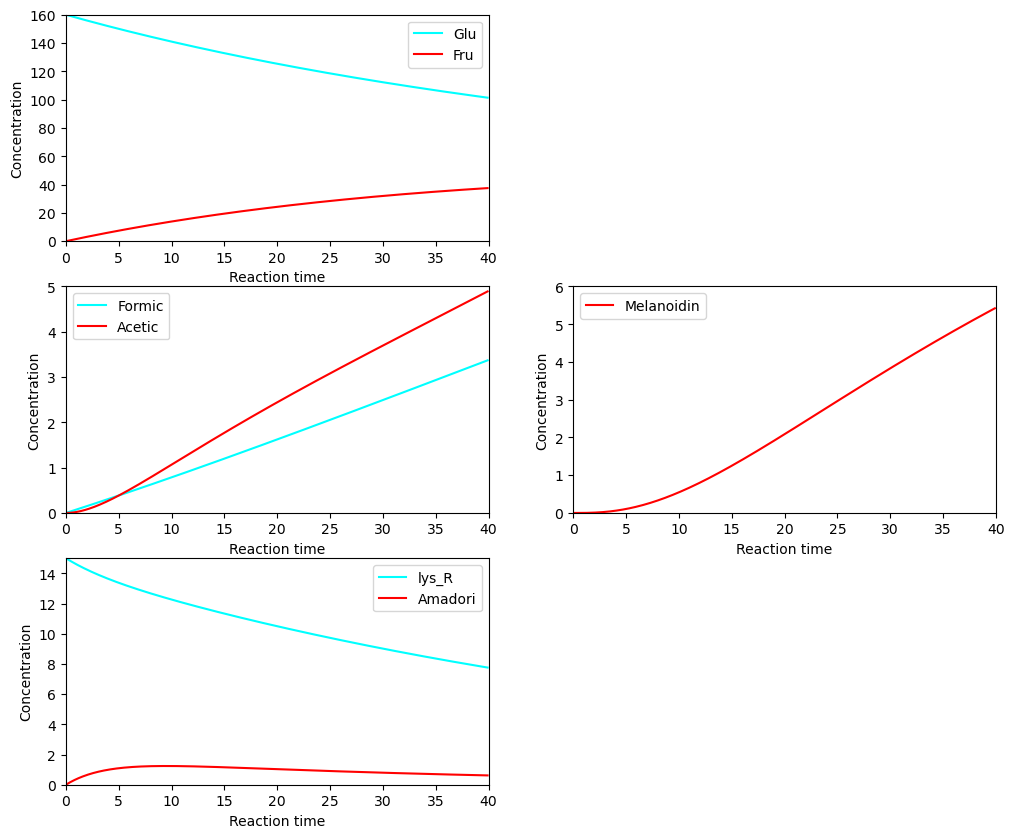
BioMD 52#
See https://www.ebi.ac.uk/biomodels/BIOMD0000000052#Curation
# load and simulate model
model, _, _, _ = load_biomodel(52)
n_secs = 40
n_steps = int(n_secs / model.deltaT)
ys, ws, ts = model(n_steps)
# plot time course simulation as in original paper
y_indexes = model.modelstepfunc.y_indexes
fig, ax = plt.subplots(3, 2, figsize=(12, 10))
ax[0,0].plot(ts, ys[y_indexes["Glu"]], color="cyan", label="Glu")
ax[0,0].plot(ts, ys[y_indexes["Fru"]], color="red", label="Fru")
ax[0,0].set_ylim([0, 160])
ax[1,0].plot(ts, ys[y_indexes["Formic_acid"]], color="cyan", label="Formic")
ax[1,0].plot(ts, ys[y_indexes["Acetic_acid"]], color="red", label="Acetic")
ax[1,0].set_ylim([0, 5])
ax[2,0].plot(ts, ys[y_indexes["lys_R"]], color="cyan", label="lys_R")
ax[2,0].plot(ts, ys[y_indexes["Amadori"]], color="red", label="Amadori")
ax[2,0].set_ylim([0, 15])
ax[1,1].plot(ts, ys[y_indexes["Melanoidin"]], color="red", label="Melanoidin")
ax[1,1].set_ylim([0, 6])
for i in range(3):
for j in range(2):
if (i, j) not in [(0,1), (2, 1)]:
ax[i,j].set_xlim([0,40])
ax[i,j].set_xlabel("Reaction time")
ax[i,j].set_ylabel("Concentration")
ax[i,j].legend()
else:
ax[i,j].axis("off")
plt.show()
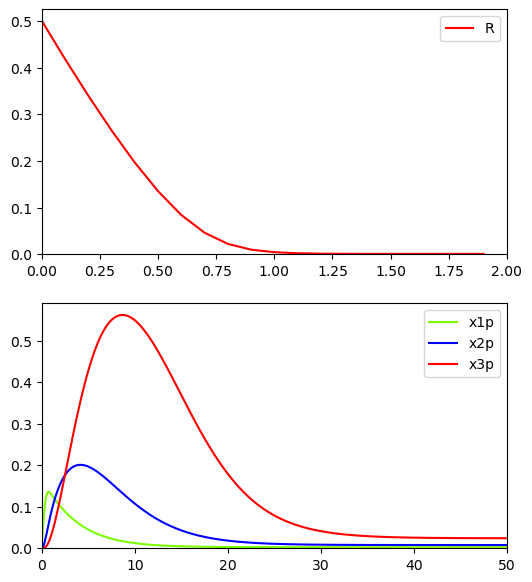
BioMD 84#
See https://www.ebi.ac.uk/biomodels/BIOMD0000000084#Curation
# load and simulate model
model, _, _, _ = load_biomodel(84)
n_secs = 50
n_steps = int(n_secs / model.deltaT)
ys, ws, ts = model(n_steps)
# plot time course simulation as in original paper
y_indexes = model.modelstepfunc.y_indexes
fig, ax = plt.subplots(2, 1, figsize=(6, 7))
ax[0].plot(
ts[: int(2 / model.deltaT)],
ys[y_indexes["R"], : int(2 / model.deltaT)],
color="red",
label="R",
)
ax[0].legend()
ax[0].set_xlim([0.,2])
ax[0].set_ylim(bottom=0)
ax[1].plot(ts, ys[y_indexes["x1p"]], color="lawngreen", label="x1p")
ax[1].plot(ts, ys[y_indexes["x2p"]], color="blue", label="x2p")
ax[1].plot(ts, ys[y_indexes["x3p"]], color="red", label="x3p")
ax[1].legend()
ax[1].set_xlim([0.,50])
ax[1].set_ylim(bottom=0)
plt.show()
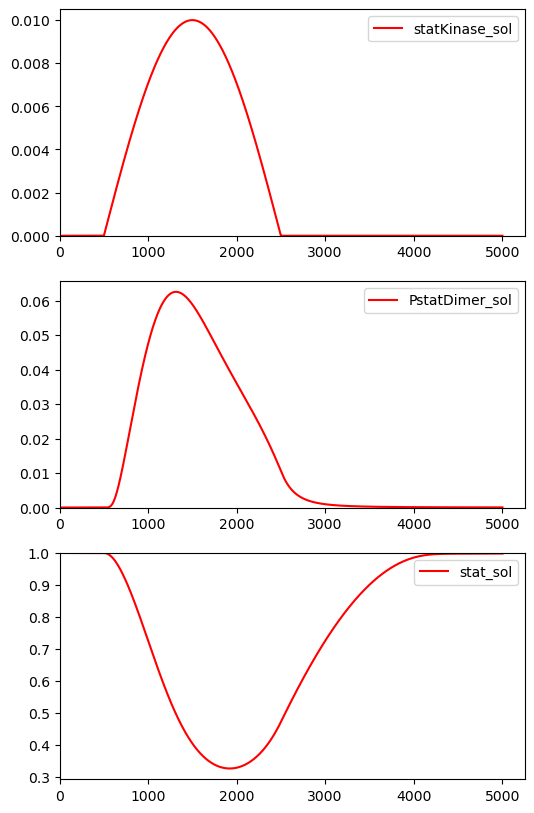
BioMD 167#
See https://www.ebi.ac.uk/biomodels/BIOMD0000000167#Curation
# load and simulate model
model, _, _, _ = load_biomodel(167)
n_secs = 5000
n_steps = int(n_secs / model.deltaT)
ys, ws, ts = model(n_steps)
# plot time course simulation as in original paper
w_indexes = model.modelstepfunc.w_indexes
y_indexes = model.modelstepfunc.y_indexes
fig, ax = plt.subplots(3, 1, figsize=(6, 10))
ax[0].plot(ts, ws[w_indexes["statKinase_sol"]]/14.625, color="red", label="statKinase_sol")
ax[0].legend()
ax[1].plot(ts, ys[y_indexes["PstatDimer_sol"]]/14.625, color="red", label="PstatDimer_sol")
ax[1].legend()
ax[2].plot(ts, ys[y_indexes["stat_sol"]]/14.625, color="red", label="stat_sol")
ax[2].legend()
for i in range(3):
ax[i].set_xlim(left=0)
if i<2:
ax[i].set_ylim(bottom=0)
else:
ax[i].set_ylim(top=1)
plt.show()
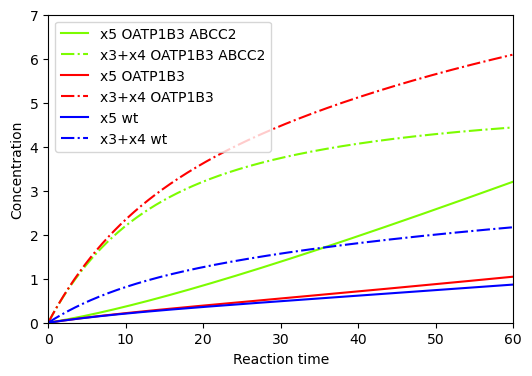
BioMD 197#
See https://www.ebi.ac.uk/biomodels/BIOMD0000000197#Curation
# load and simulate model
model, _, _, c = load_biomodel(197)
n_secs = 60
n_steps = int(n_secs / model.deltaT)
# sim default
ys, _, ts = model(n_steps)
# sim 1
c_OATP1B3 = c.at[1].set(0.)
ys_OATP1B3, _, ts = model(n_steps, c=c_OATP1B3)
# sim 2
c_wt = c.at[:2].set(0.)
ys_wt, _, ts = model(n_steps, c=c_wt)
# plot time course simulation as in original paper
y_indexes = model.modelstepfunc.y_indexes
plt.figure(figsize=(6, 4))
plt.plot(ts, ys[y_indexes["x5"]], color="lawngreen", label="x5 OATP1B3 ABCC2")
plt.plot(ts, ys[y_indexes["x3"]]+ys[y_indexes["x4"]], color="lawngreen", linestyle="-.", label="x3+x4 OATP1B3 ABCC2")
plt.plot(ts, ys_OATP1B3[y_indexes["x5"]], color="red", label="x5 OATP1B3")
plt.plot(ts, ys_OATP1B3[y_indexes["x3"]]+ys_OATP1B3[y_indexes["x4"]], color="red", linestyle="-.", label="x3+x4 OATP1B3")
plt.plot(ts, ys_wt[y_indexes["x5"]], color="blue", label="x5 wt")
plt.plot(ts, ys_wt[y_indexes["x3"]]+ys_wt[y_indexes["x4"]], color="blue", linestyle="-.", label="x3+x4 wt")
plt.xlabel("Reaction time")
plt.ylabel("Concentration")
plt.xlim([0,60])
plt.ylim([0,7])
plt.legend()
plt.show()
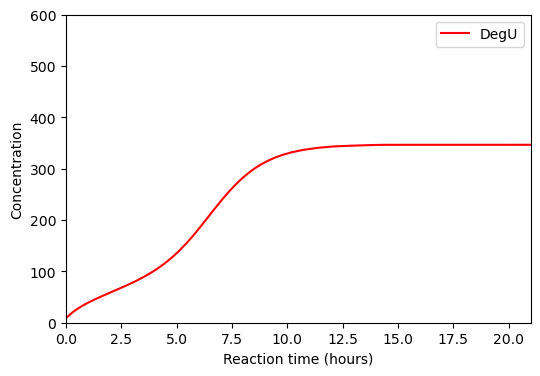
BioMD 240#
See https://www.ebi.ac.uk/biomodels/BIOMD0000000240#Curation
# load and simulate model
model, _, _, c = load_biomodel(240)
n_secs = 21*3600
n_steps = int(n_secs / model.deltaT)
ys, ws, ts = model(n_steps)
# plot time course simulation as in original paper
w_indexes = model.modelstepfunc.w_indexes
plt.figure(figsize=(6, 4))
plt.plot(ts/3600, ws[w_indexes["DegU_Total"]], color="red", label="DegU")
plt.xlabel("Reaction time (hours)")
plt.ylabel("Concentration")
plt.xlim([0,21])
plt.ylim([0,600])
plt.legend()
plt.show()
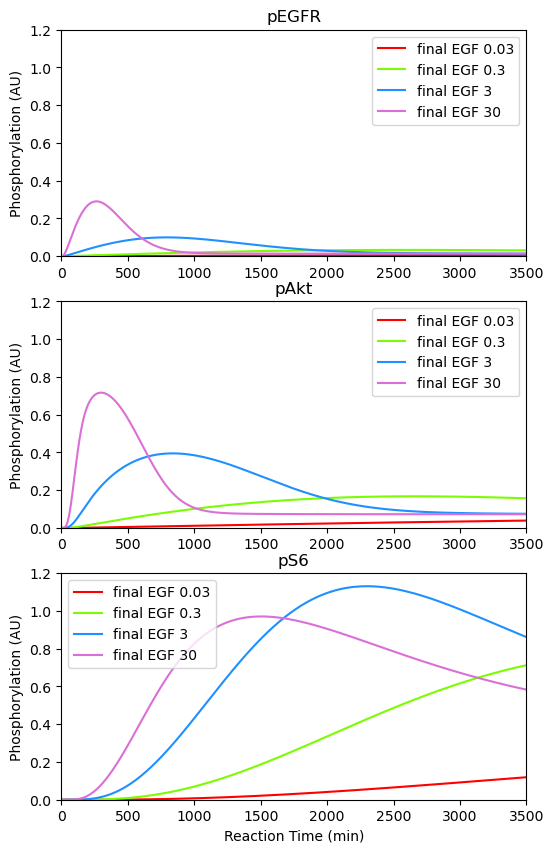
BioMD 262#
See https://www.ebi.ac.uk/biomodels/BIOMD0000000262#Curation
# load and simulate model
model, _, _, c = load_biomodel(262)
c_indexes = model.modelstepfunc.c_indexes
w_indexes = model.modelstepfunc.w_indexes
y_indexes = model.modelstepfunc.y_indexes
n_secs = 3500
n_steps = int(n_secs / model.deltaT)
# simulations
c = c.at[c_indexes['EGF_conc_ramp']].set(.03)
ys_1, ws_1, ts = model(n_steps, c=c)
c = c.at[c_indexes['EGF_conc_ramp']].set(.3)
ys_2, ws_2, ts = model(n_steps, c=c)
c = c.at[c_indexes['EGF_conc_ramp']].set(3)
ys_3, ws_3, ts = model(n_steps, c=c)
c = c.at[c_indexes['EGF_conc_ramp']].set(30)
ys_4, ws_4, ts = model(n_steps, c=c)
# plot time course simulation as in original paper
fig, ax = plt.subplots(3, 1, figsize=(6, 10))
ax[0].plot(ts, ys_1[y_indexes["pEGFR"]]*c[c_indexes["pEGFR_scaleFactor"]], color="red", label="final EGF 0.03")
ax[0].plot(ts, ys_2[y_indexes["pEGFR"]]*c[c_indexes["pEGFR_scaleFactor"]], color="lawngreen", label="final EGF 0.3")
ax[0].plot(ts, ys_3[y_indexes["pEGFR"]]*c[c_indexes["pEGFR_scaleFactor"]], color="dodgerblue", label="final EGF 3")
ax[0].plot(ts, ys_4[y_indexes["pEGFR"]]*c[c_indexes["pEGFR_scaleFactor"]], color="orchid", label="final EGF 30")
ax[0].set_title("pEGFR")
ax[1].plot(ts, ys_1[y_indexes["pAkt"]]*c[c_indexes["pAkt_scaleFactor"]], color="red", label="final EGF 0.03")
ax[1].plot(ts, ys_2[y_indexes["pAkt"]]*c[c_indexes["pAkt_scaleFactor"]], color="lawngreen", label="final EGF 0.3")
ax[1].plot(ts, ys_3[y_indexes["pAkt"]]*c[c_indexes["pAkt_scaleFactor"]], color="dodgerblue", label="final EGF 3")
ax[1].plot(ts, ys_4[y_indexes["pAkt"]]*c[c_indexes["pAkt_scaleFactor"]], color="orchid", label="final EGF 30")
ax[1].set_title("pAkt")
ax[2].plot(ts, ys_1[y_indexes["pS6"]]*c[c_indexes["pS6_scaleFactor"]], color="red", label="final EGF 0.03")
ax[2].plot(ts, ys_2[y_indexes["pS6"]]*c[c_indexes["pS6_scaleFactor"]], color="lawngreen", label="final EGF 0.3")
ax[2].plot(ts, ys_3[y_indexes["pS6"]]*c[c_indexes["pS6_scaleFactor"]], color="dodgerblue", label="final EGF 3")
ax[2].plot(ts, ys_4[y_indexes["pS6"]]*c[c_indexes["pS6_scaleFactor"]], color="orchid", label="final EGF 30")
ax[2].set_title("pS6")
ax[2].set_xlabel("Reaction Time (min)")
for i in range(3):
ax[i].set_xlim([0, 3500])
ax[i].set_ylim([0, 1.2])
ax[i].legend()
ax[i].set_ylabel("Phosphorylation (AU)")
plt.show()
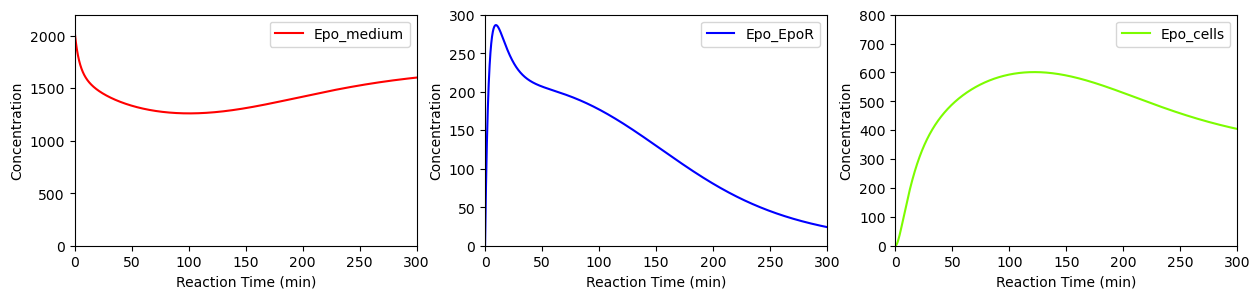
BioMD 271#
See https://www.ebi.ac.uk/biomodels/BIOMD0000000271#Curation
# load and simulate model
model, _, _, c = load_biomodel(271)
w_indexes = model.modelstepfunc.w_indexes
y_indexes = model.modelstepfunc.y_indexes
n_secs = 300
n_steps = int(n_secs / model.deltaT)
# simulation
ys, ws, ts = model(n_steps, c=c)
# plot time course simulation as in original paper
fig, ax = plt.subplots(1, 3, figsize=(15, 3))
ax[0].plot(ts, ws[w_indexes["Epo_medium"]], color="red", label="Epo_medium")
ax[0].set_ylim([0,2200])
ax[1].plot(ts, ys[y_indexes["Epo_EpoR"]], color="blue", label="Epo_EpoR")
ax[1].set_ylim([0,300])
ax[2].plot(ts, ws[w_indexes["Epo_cells"]], color="lawngreen", label="Epo_cells")
ax[2].set_ylim([0,800])
for i in range(3):
ax[i].set_xlim([0,300])
ax[i].legend()
ax[i].set_ylabel("Concentration")
ax[i].set_xlabel("Reaction Time (min)")
plt.show()
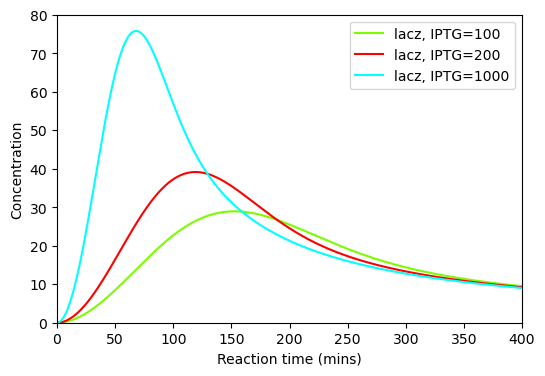
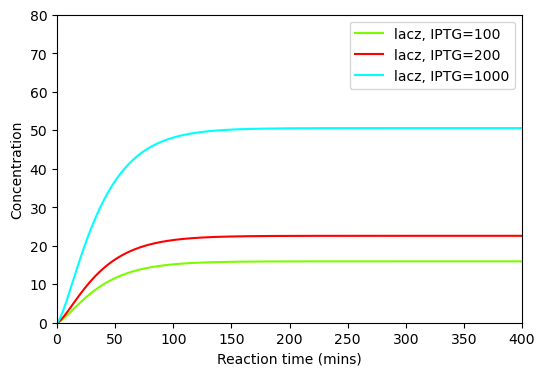
BioMD 459#
See https://www.ebi.ac.uk/biomodels/BIOMD0000000459#Curation
# load model
model, _, _, c = load_biomodel(459)
n_secs = 400
n_steps = int(n_secs / model.deltaT)
y_indexes = model.modelstepfunc.y_indexes
c_indexes = model.modelstepfunc.c_indexes
# simulate model
c = c.at[c_indexes['IPTG']].set(100.)
ys_100, _, ts = model(n_steps, c=c)
c = c.at[c_indexes['IPTG']].set(200.)
ys_200, _, ts = model(n_steps, c=c)
c = c.at[c_indexes['IPTG']].set(1000.)
ys_1000, _, ts = model(n_steps, c=c)
# plot time course simulation as in original paper
plt.figure(figsize=(6, 4))
plt.plot(ts, ys_100[y_indexes["lacz"]], color="lawngreen", label="lacz, IPTG=100")
plt.plot(ts, ys_200[y_indexes["lacz"]], color="red", label="lacz, IPTG=200")
plt.plot(ts, ys_1000[y_indexes["lacz"]], color="cyan", label="lacz, IPTG=1000")
plt.xlabel("Reaction time (mins)")
plt.ylabel("Concentration")
plt.xlim([0,400])
plt.ylim([0,80])
plt.legend()
plt.show()
BioMD 461#
See https://www.ebi.ac.uk/biomodels/BIOMD0000000461#Curation
# load model
model, _, _, c = load_biomodel(461)
n_secs = 400
n_steps = int(n_secs / model.deltaT)
y_indexes = model.modelstepfunc.y_indexes
c_indexes = model.modelstepfunc.c_indexes
# simulate model
c = c.at[c_indexes['IPTG']].set(100.)
ys_100, _, ts = model(n_steps, c=c)
c = c.at[c_indexes['IPTG']].set(200.)
ys_200, _, ts = model(n_steps, c=c)
c = c.at[c_indexes['IPTG']].set(1000.)
ys_1000, _, ts = model(n_steps, c=c)
# plot time course simulation as in original paper
plt.figure(figsize=(6, 4))
plt.plot(ts, ys_100[y_indexes["lacz"]], color="lawngreen", label="lacz, IPTG=100")
plt.plot(ts, ys_200[y_indexes["lacz"]], color="red", label="lacz, IPTG=200")
plt.plot(ts, ys_1000[y_indexes["lacz"]], color="cyan", label="lacz, IPTG=1000")
plt.xlabel("Reaction time (mins)")
plt.ylabel("Concentration")
plt.xlim([0,400])
plt.ylim([0,80])
plt.legend()
plt.show()
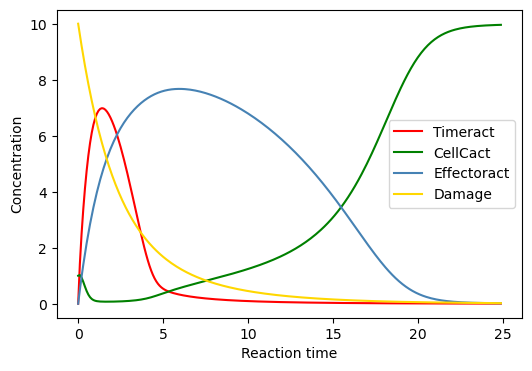
BioMD 641#
See https://www.ebi.ac.uk/biomodels/BIOMD0000000641#Curation
# load and simulate model
model, _, _, c = load_biomodel(641)
n_secs = 25
n_steps = int(n_secs / model.deltaT)
ys, ws, ts = model(n_steps)
# plot time course simulation as in original paper
y_indexes = model.modelstepfunc.y_indexes
w_indexes = model.modelstepfunc.w_indexes
plt.figure(figsize=(6, 4))
plt.plot(ts, ys[y_indexes["Timeract"]], color="red", label="Timeract")
plt.plot(ts, ys[y_indexes["CellCact"]], color="green", label="CellCact")
plt.plot(ts, ys[y_indexes["Effectoract"]], color="steelblue", label="Effectoract")
plt.plot(ts, ws[w_indexes["Damage"]], color="gold", label="Damage")
plt.xlabel("Reaction time")
plt.ylabel("Concentration")
plt.legend()
plt.show()
⚠️ Error Cases#
Note that SBMLtoODEjax does not (yet) handles the numerical simulation of all models present on the BioModels website, or more generally it does not handle all possible SBML files. We detail below the error types we obtain when trying to simulate the list of currently-hosted curated models (1048 models in total):